Platform for Targeted Precision Proteome Profiling
What are proteomes and proteome profiles?
The proteome is defined as the totality of all proteins encoded in the genes of a living being. In humans, the proteome comprises at least 20,000 unmodified ("canonical") proteins. Under normal physiological conditions only those proteins of the entire proteome are expressed that are necessary for the execution of organ functions at a given time - thus resulting in organ-specific proteome profiles. Since these profiles map the physiological state of the respective organ on a molecular level, the proteome profile of an organ changes in the course of the development and progression of a disease, even before changes in organ function can be medically detected. Some of these proteins enter the blood stream and thus make it possible to detect the course of a disease in the blood.Analysis of the Proteome
In order to gain insight into the molecular mechanisms of a disease, these changes must be depicted as comprehensively as possible. The proteome research laboratory is equipped with a high-throughput analysis platform based on immuno-qPCR technology. This technology enables the targeted, highly specific and highly sensitive simultaneous quantitative measurement of 92 proteins in 88 samples. These 92 proteins are grouped into a panel. Currently, 14 of these panels are available from Olink Proteomics (Uppsala, Sweden), covering 1,288 human proteins. These are divided into 7 panels focused on specific disease areas (2 x cardiology, 3 x oncology, 2 x neurology) and 7 panels focused on important biological processes (inflammation, immune response, cell regulation, metabolism, cardiometabolism, development, organ damage). In addition, one panel of 92 mouse proteins is available. The measurable sample qualities mainly include blood plasma and serum, but also urine, cerebrospinal fluid, saliva, synovial fluids and tissue lysates.The proteome research laboratory has been certified by Olink Proteomics to carry out the analyses and is registered as Shared Expertise (SE175) with the German Centre for Cardiovascular Research e.V. (https://dzhk.de/forschung/praeklinische-forschung/shared-expertise/).
The measurement of several of these panels has already enabled the identification of protein biomarkers in the areas of venous thromboembolic events, heart failure and type 2 diabetes mellitus.
Technology
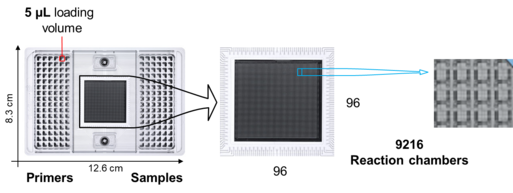
The protein measurements are carried out on Dynamic Array chips from Fluidigm (Figure 1) with integrated fluidic circuits (IFC). The measurement on these chips makes it possible to determine the proteins in an initial sample quantity of only one microlitre (0.001 millilitres).
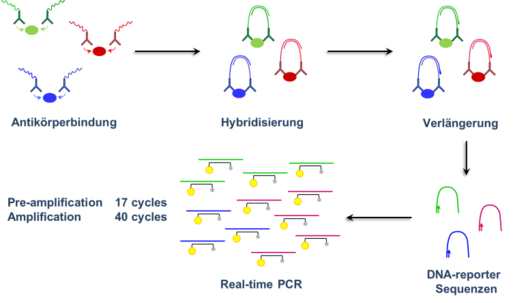
The extraordinarily high specificity of immuno-qPCR-based protein determinations results from the need for spatial proximity binding of 2 different antibodies to the same molecule of a target protein ® Olink Proximity Extension Assay (PEA) technology (Figure 2). Both antibodies are chemically linked to complementary oligonucleotides that hybridise due to proximity. Only when hybridisation is complete, protein-specific DNA reporter sequences are formed by enzymatic extension of the oligonucleotides, which are detected by amplification using qPCR. This amplification of the reporter sequences results in the extraordinarily high sensitivity of the determination. The resulting signal is proportional to the content of the respective protein in the sample.
Contact

Dr. Thomas Köck
Co-Head of Research Laboratory | BioBank Mainz
Back to Scientific Expertise